12/24/24–SciMETv3 Single-cell Atlas paper released in Cell Genomics: Atlas-scale Single-cell DNA methylation profiling with SciMETv3 Ruth v Nichols, Lauren E. Rylaarsdam, Brendan L. O’connell, Zohar Shipony, Nika Iremadze, Sonia N. Acharya, Andrew Adey
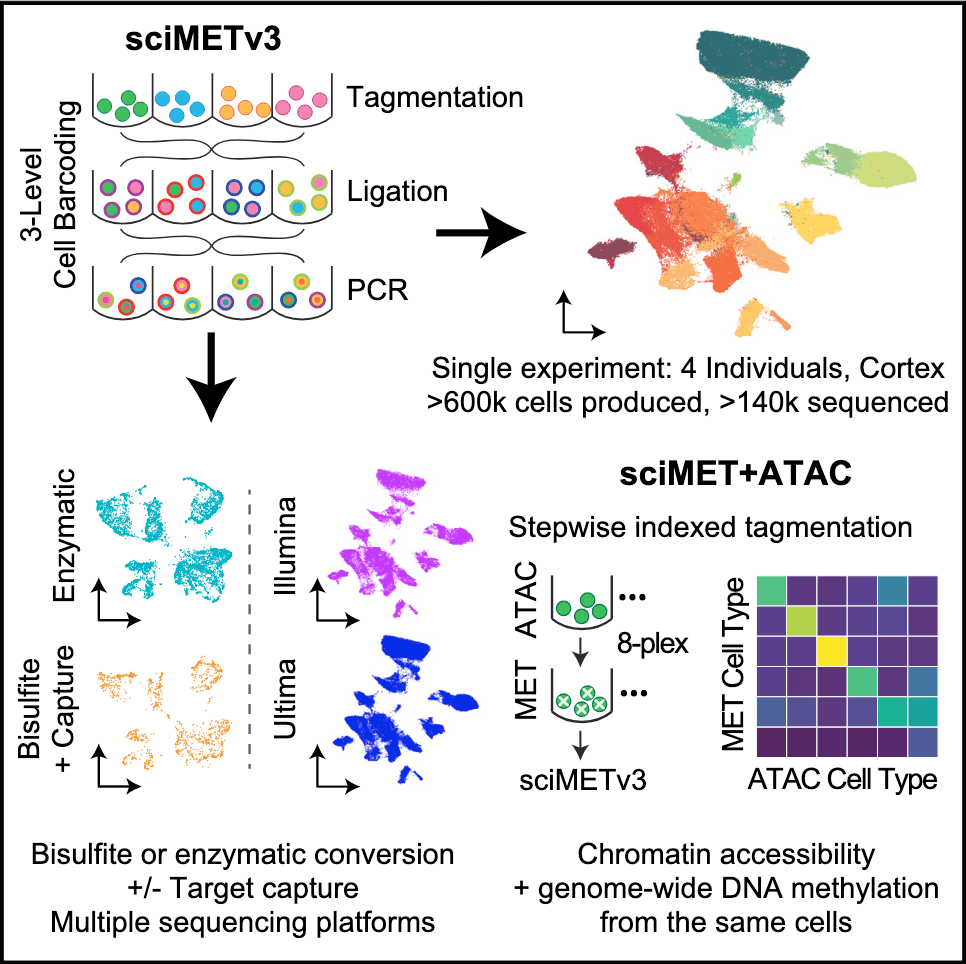
8/30/24– SciMETv3 Preprint Released in BioRxiv: Atlas-scale Single-cell DNA methylation profiling with SciMETv3
08/27/2024– New Preprint in BioRxiv: Single-cell DNA methylation analysis tool Amethyst reveals distinct noncanonical methylation patterns in human glial cells
Github link: An analysis toolkit for single-cell methylation sequencing data
lrylaarsdam/Amethyst
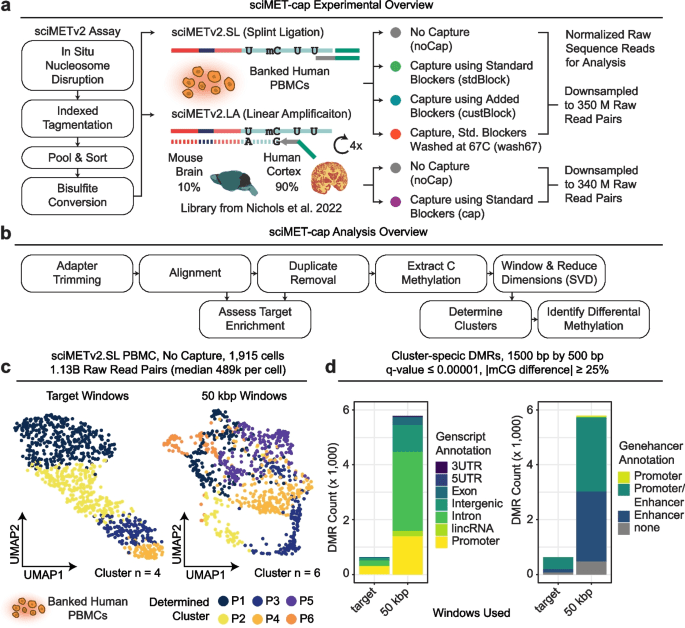
7/10/2024– Adey Lab SciMET-cap paper Just released in Genome Biology: SciMET-cap: high-throughput single-cell methylation analysis with reduced sequencing burden Sonia N. Acharya, Ruth V. Nichols, Lauren E. Rylaarsdam, Brendan L. O’Connell, Theodore P. Braun and Andrew C. Adey.
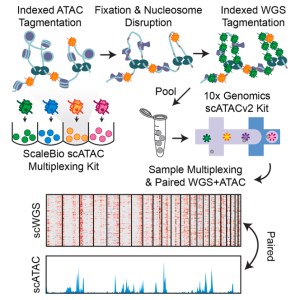
6/10/24– We’d like to highlight a recent publication in which our Graduate Student, Konstantin Queitsch, is first author : Accessible high-throughput single-cell whole-genome sequencing with paired chromatin accessibility. Now in Cell Reports Methods.
Here is a summary figure below for the paper:
4/18/24- Shown below are the models of the developing brain called “Cerebral Organoids” grown in culture by our postdoc, Lauren Rylaarsdam.
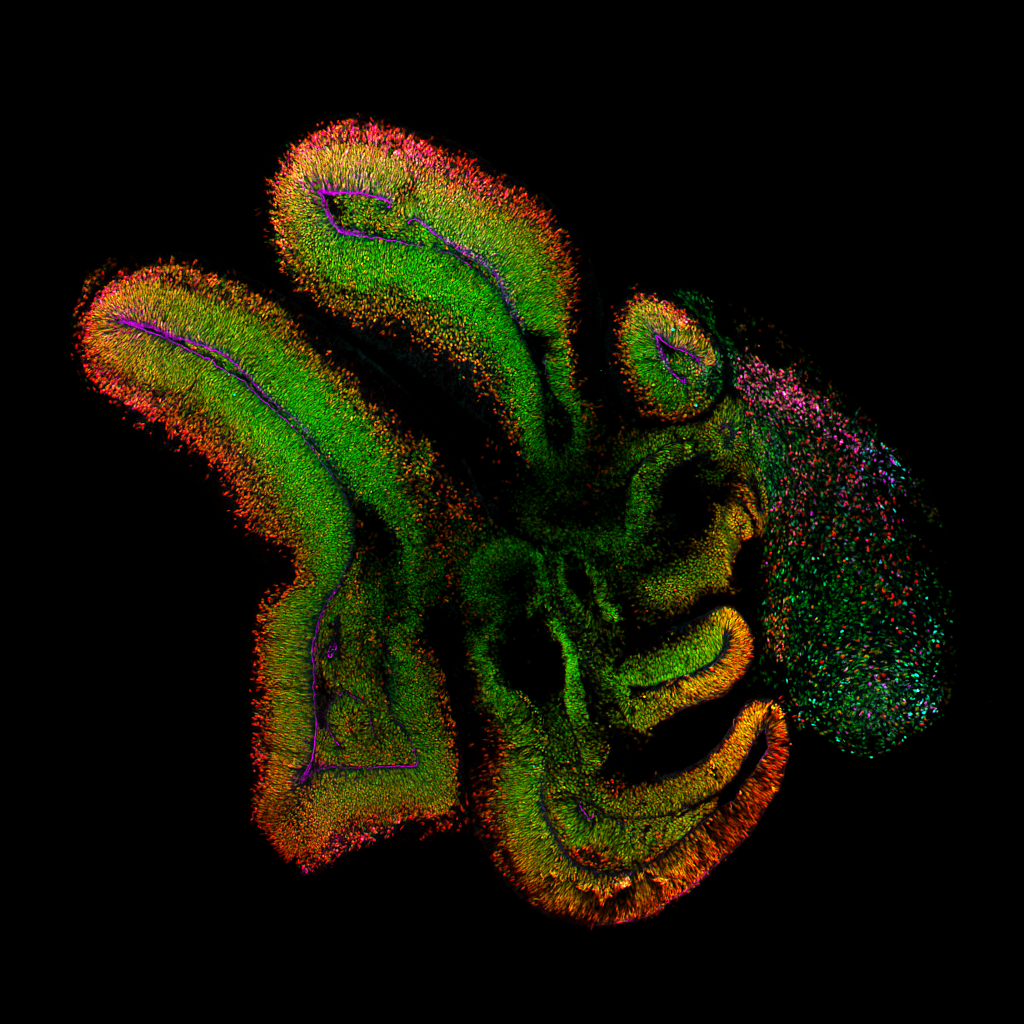
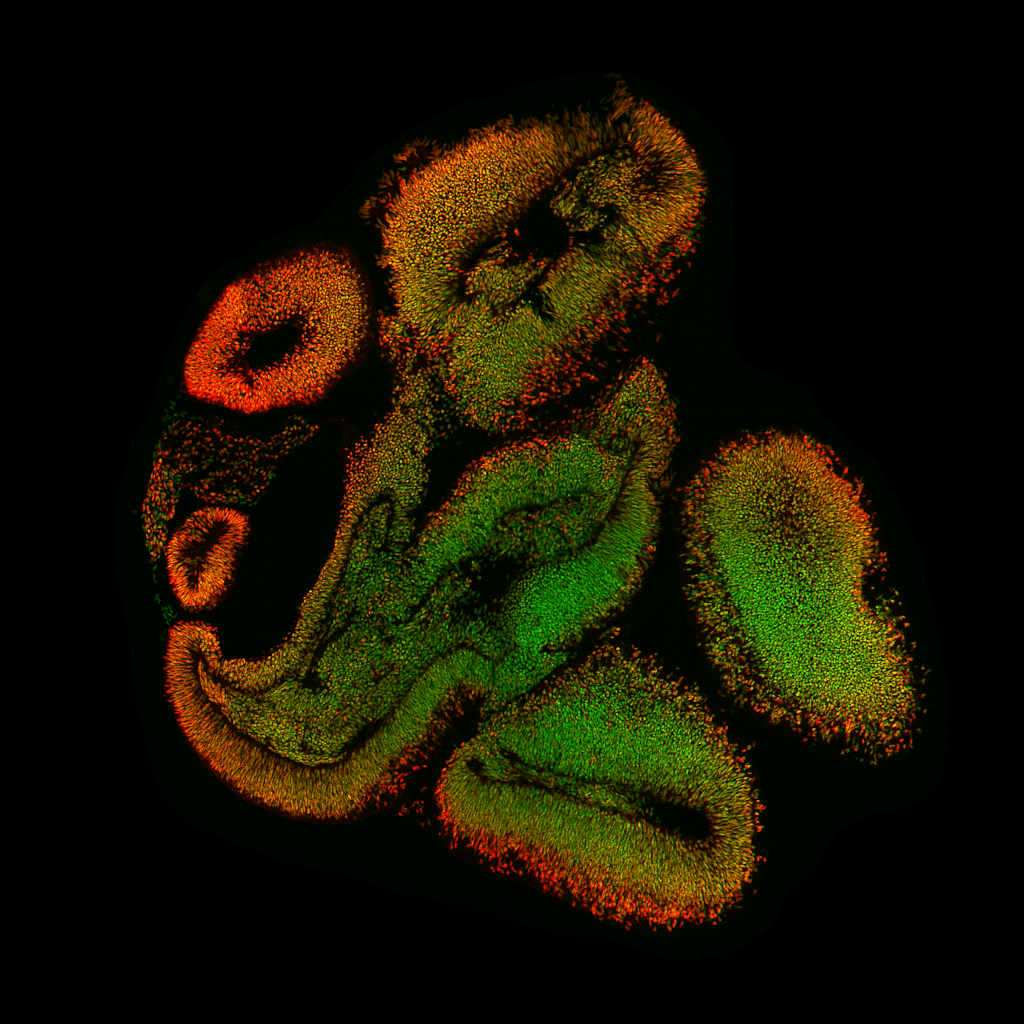
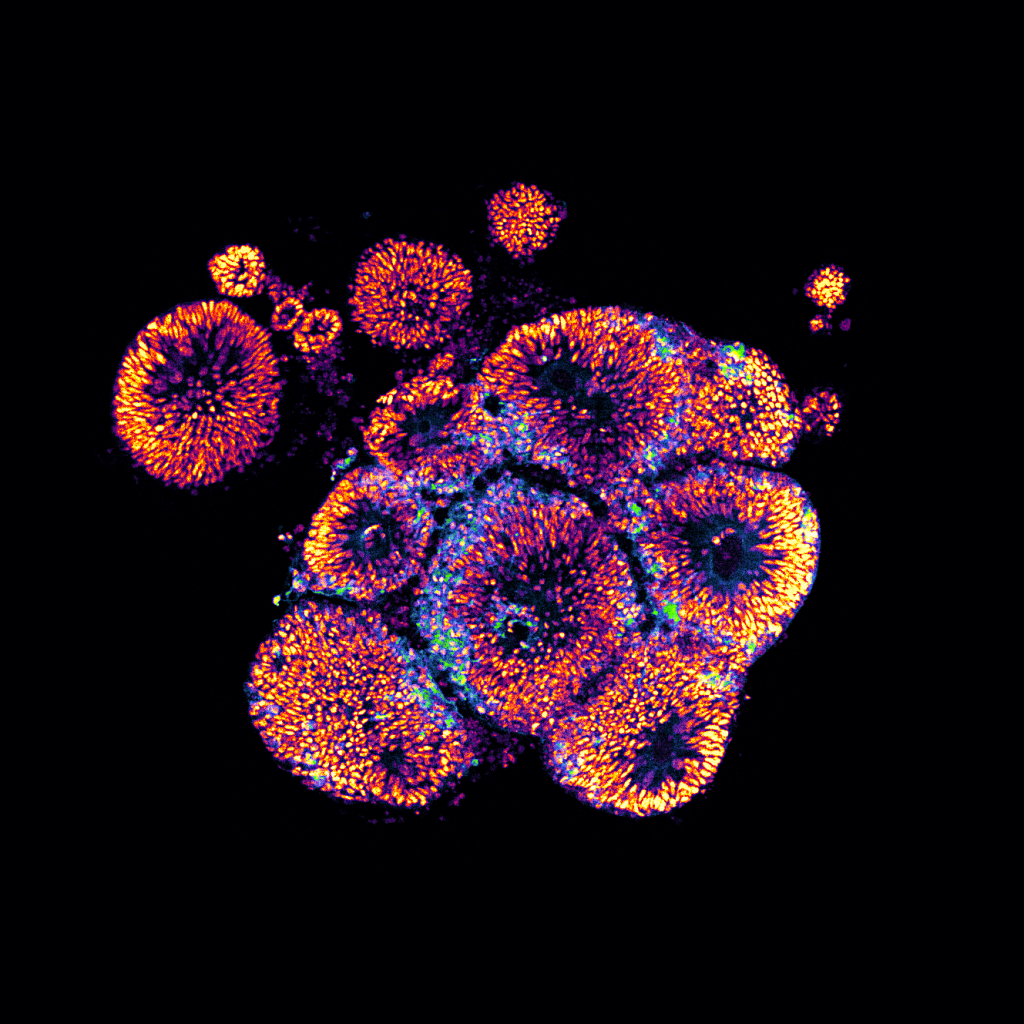
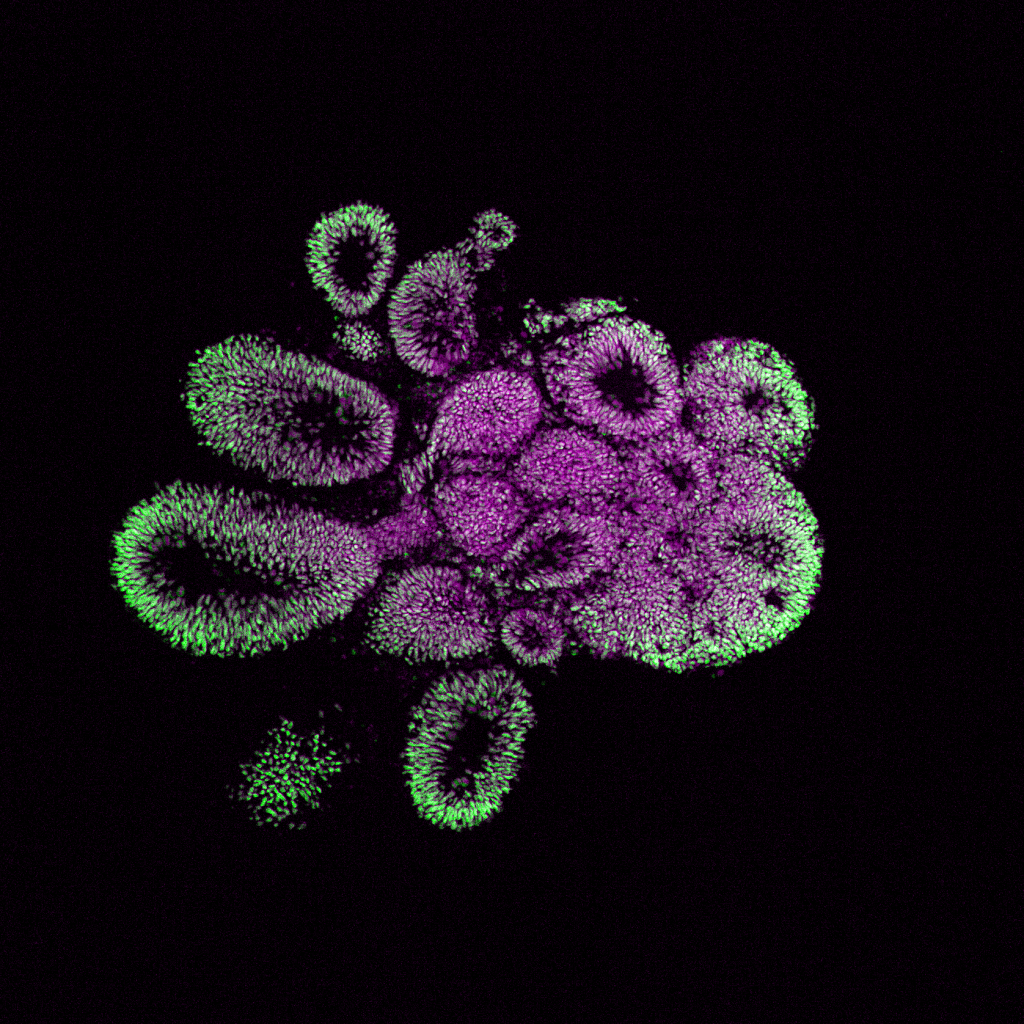
Cerebral Organoids are spheres of pluripotent cells exposed to a series of growth factors in culture that direct them towards a neuronal identity. These organoids reproduce many aspects of the developing human brain, yielding valuable insights into an otherwise inaccessible process.
7/14/23 – Lots of updates!! First off, we have two new publications and two new preprints (details below); Andrew Adey was promoted to full Professor with Tenure; and we have a new postdoc in the lab: Dr. Lauren Rylaarsdam who will be leading exciting work in the neuroscience space! Updates to the personnel tabs will come eventually.
High-throughput robust single-cell DNA methylation profiling with sciMETv2. which came out in Nature Communications
Atlas-scale single-cell chromatin accessibility using nanowell-based combinatorial indexing. in Genome Research
txci-ATAC-seq, a massive-scale single-cell technique to profile chromatin accessibility – preprint in bioRxiv – collaboration with the Cusanovich Lab
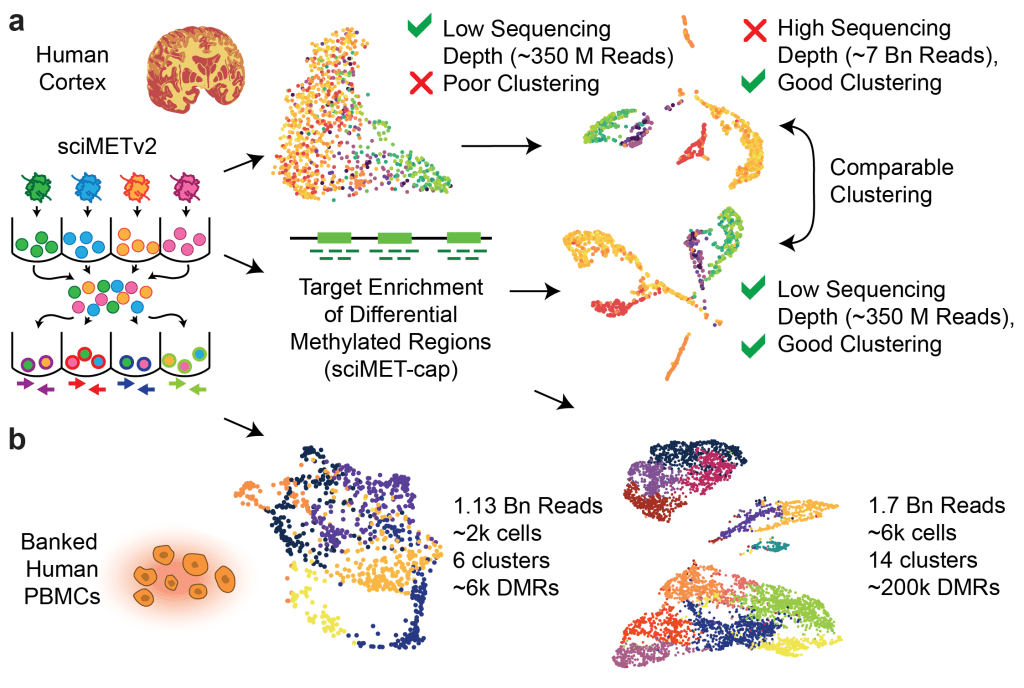
sciMET-cap: High-throughput single-cell methylation analysis with a reduced sequencing burden – preprint on bioRxiv
Below is a summary figure for the last one where we are able to drastically reduce the required sequencing depth per cell for single-cell methylation studies, bringing it much closer to the requirements for scRNA or scATAC!
12/13/22 – It has been a while since I have updated the site, but there is lots to update on! Most importantly, graduate student Casey Thornton successfully defended and was accepted into the top Laboratory Genetics and Genomics fellowship at Baylor College of Medicine! Also Fatma Erylidiz, co-mentored by Jeff Tyner also successfully defended and has taken a position back in Turkey where I am sure she will continue to lead exciting work.
We also have had some new papers – including the “s3” technology that came out in Nature Biotechnology, and most recently a new version of our sciMET technology: sciMETv2 published in Nature Communications. The new version is FAR easier to perform than v1, it does not require custom sequencing primers or recipes, is more robust, and provides way more coverage. The paper was also selected as a featured article by the journal.
2/24/2021 – Two big news items today! Graduate student Ryan Mulqueen successfully defended his Ph.D. thesis! Congratulations Dr. Mulqueen!
Our publication on the sciMAP-ATAC technology was also just published in Nature Communications! Congratulations to the lead author, graduate student Casey Thornton!
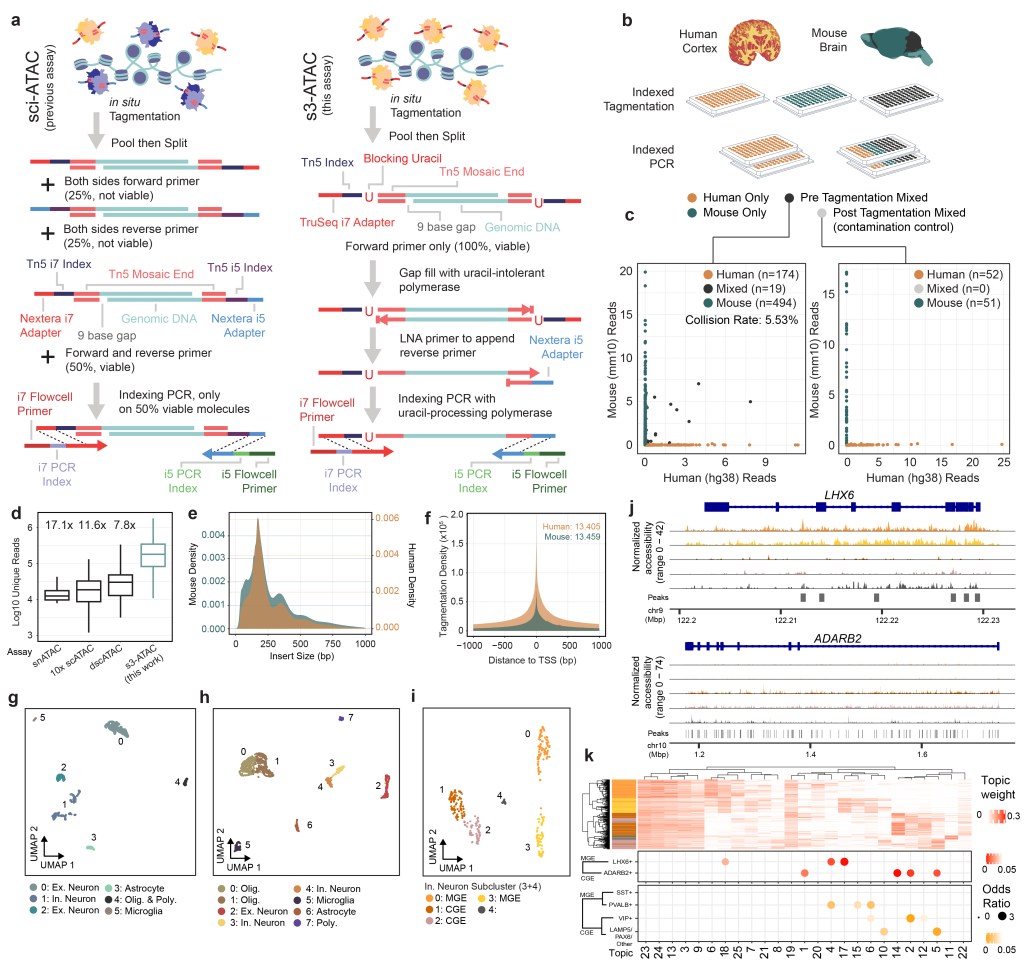
1/12/2021 – I have been slacking about updating this site, but the big news now is a new preprint! We present you with “s3” – an abbreviation for a far-too-long name: “symmetrical strand single-cell combinatorial indexing”! Graduate student Ryan Mulqueen (set to defend in February!) led work to develop this technology to improve the coverage of single-cell assays including s3-ATAC, s3-WGS, and s3-GCC (WGS+HiC)! We achieve massive boosts in unique reads per cell! Below is Figure 1!
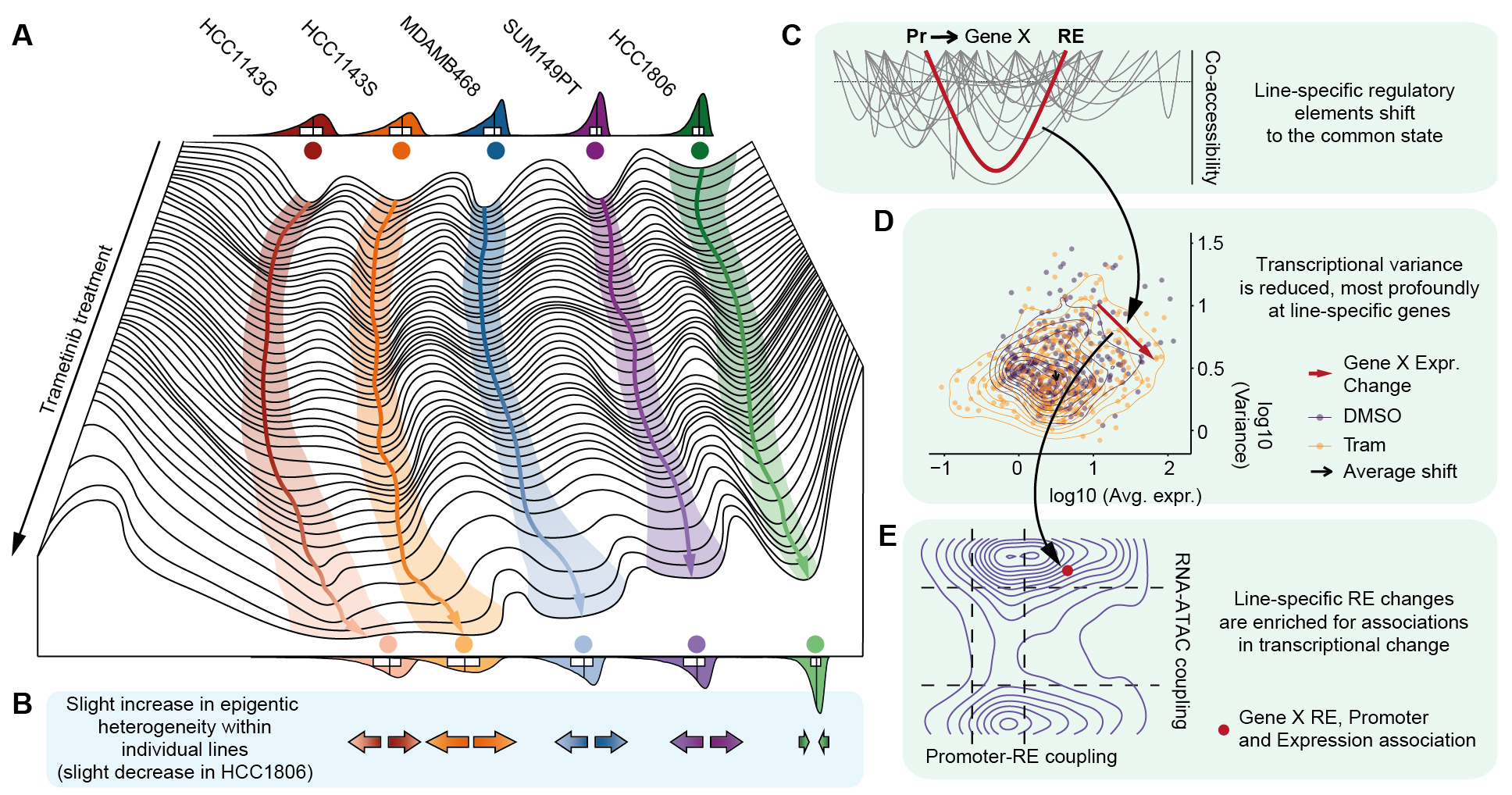
We also put out another preprint led by former graduate student Kristof Torkenczy (who is now a postdoc in the Satija Lab) on a deep analysis of chromatin and transcriptional dynamics in cancer cell lines treated with the MEK inhibitor, Trametinib. We uncover a very interesting new mechanism of cell state homogenization – give it a read to learn more! Here is a summary figure from the preprint:
Also expect to see sciMAP-ATAC (below) out in a journal soon!
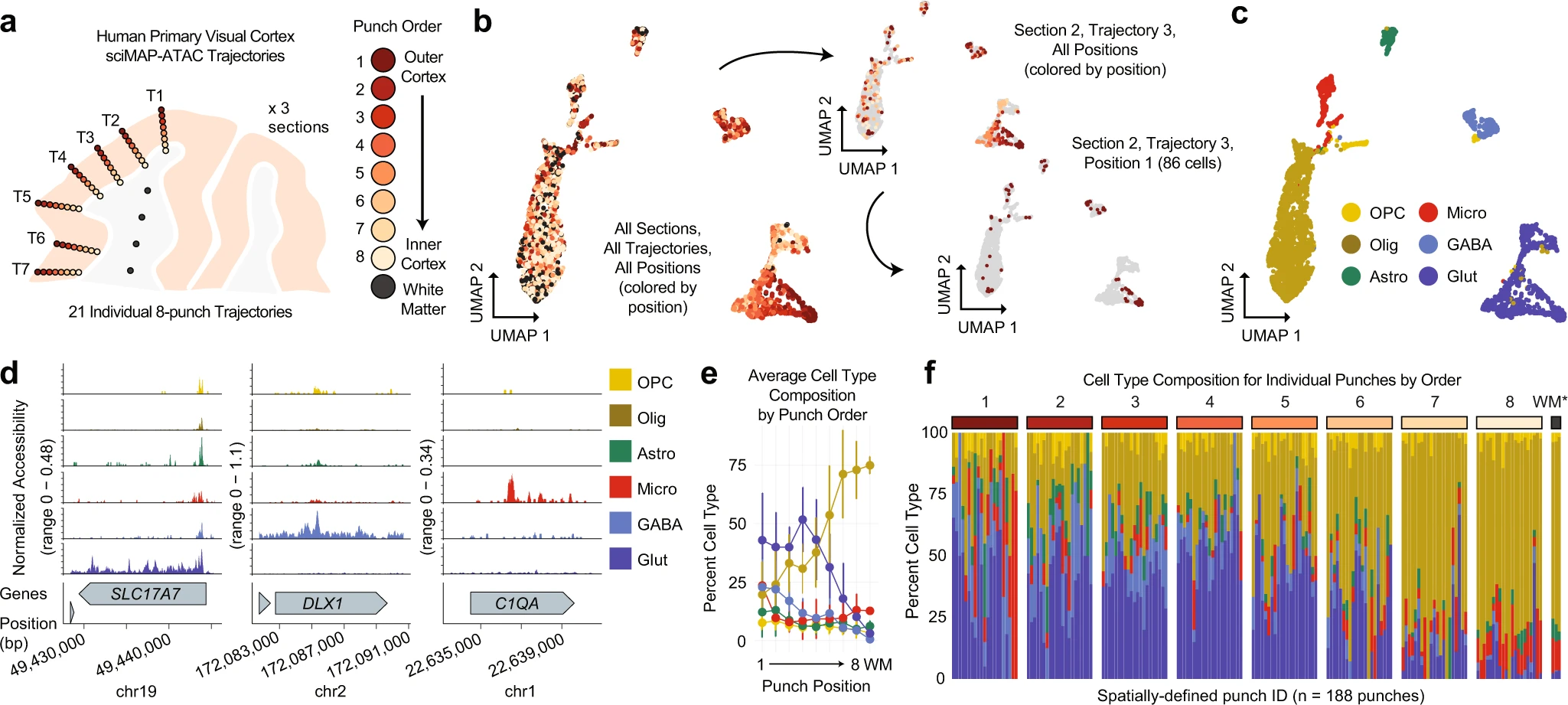
10/23/2019 – We just put out our preprint on Spatially-mapped single-cell chromatin accessibility! (sciMAP-ATAC) Led by graduate student Casey Thornton, we leverage the inherent multiplexing capabilities of our combinatorial indexing single-cell ATAC-seq workflow to enable high-density multiregional sampling. We apply it to the mouse brain with many more to come.
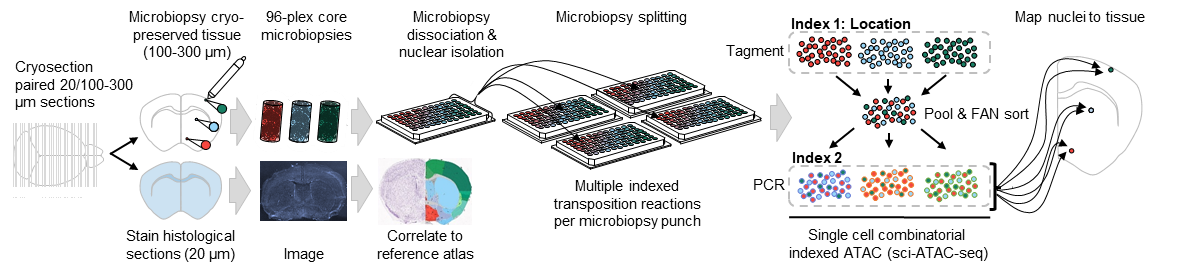
10/1/2019 – New collaborative work out in Molecular Cell led by Shendure Lab postdoc Yi Yin on a new version of sci-DNA-seq: sci-L3 that enables higher throughput, better genome coverage, target enrichment, as well as co-assays! She applied the technique to study crossovers and mis-segregation events in interspecific hybrid mice.
6/1/2019 – Dr. Adey was promoted to Associate Professor. Thank you to all of the hard working members of the lab that have enabled the success we have had!
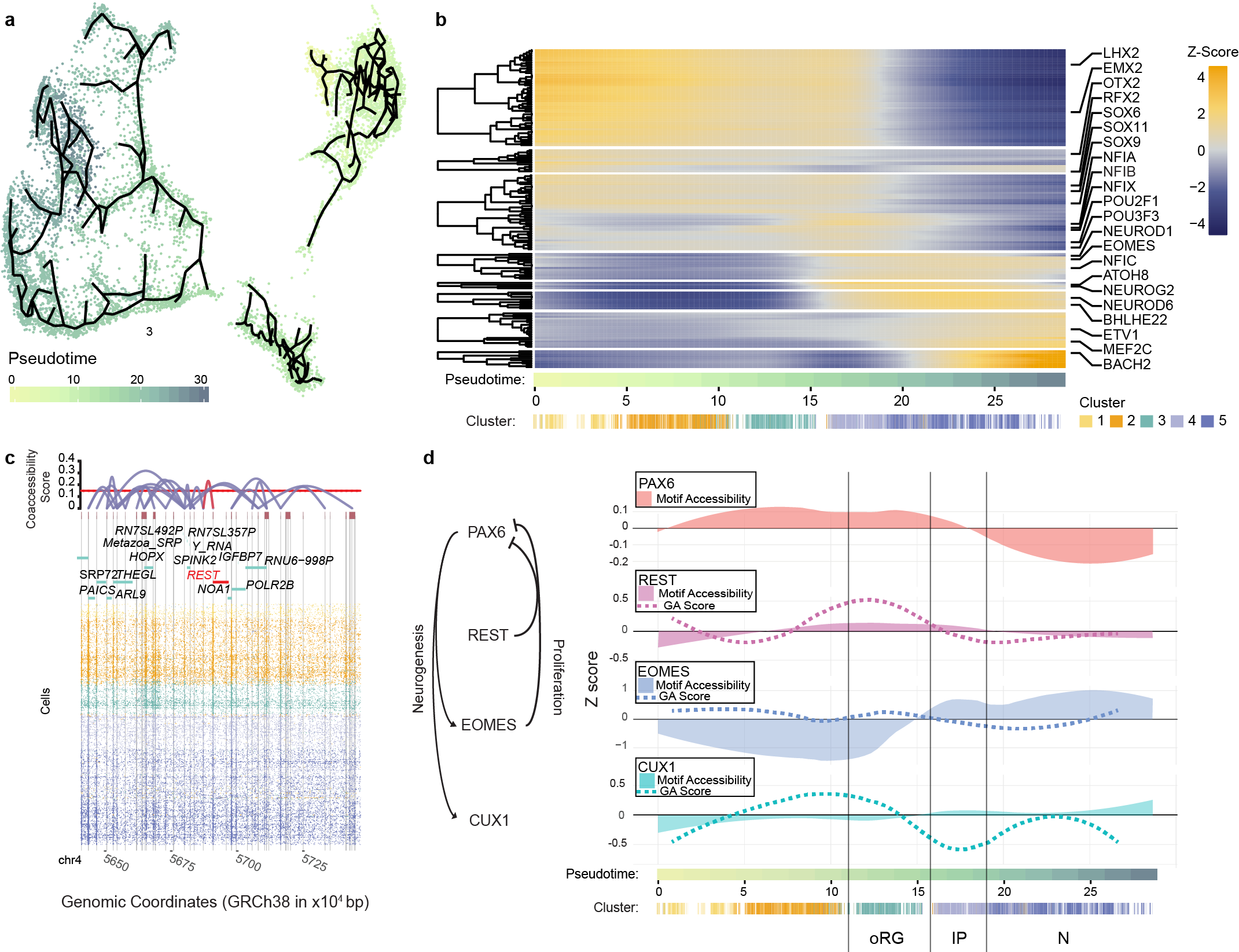
5/15/2019 – New preprint! Very excited about this collaboration with the O’Roak Lab! We use a small molecule additive during nuclei isolation and in situ tagmentation in our single-cell ATAC-seq workflow that boosts entry into the nucleus and subsequently the number of unique reads obtained per cell. We then apply it to an iPSC cortical organoid model to explore the chromatin dynamics of corticogenesis. Great work led by Ryan Mulqueen! Below is Figure 3 showing pseudotemporal ordering of cells during differentiation.
4/1/2019 – Our paper on using single-cell ATAC-seq to profile the murine hippocampus just came out! Check it out here!
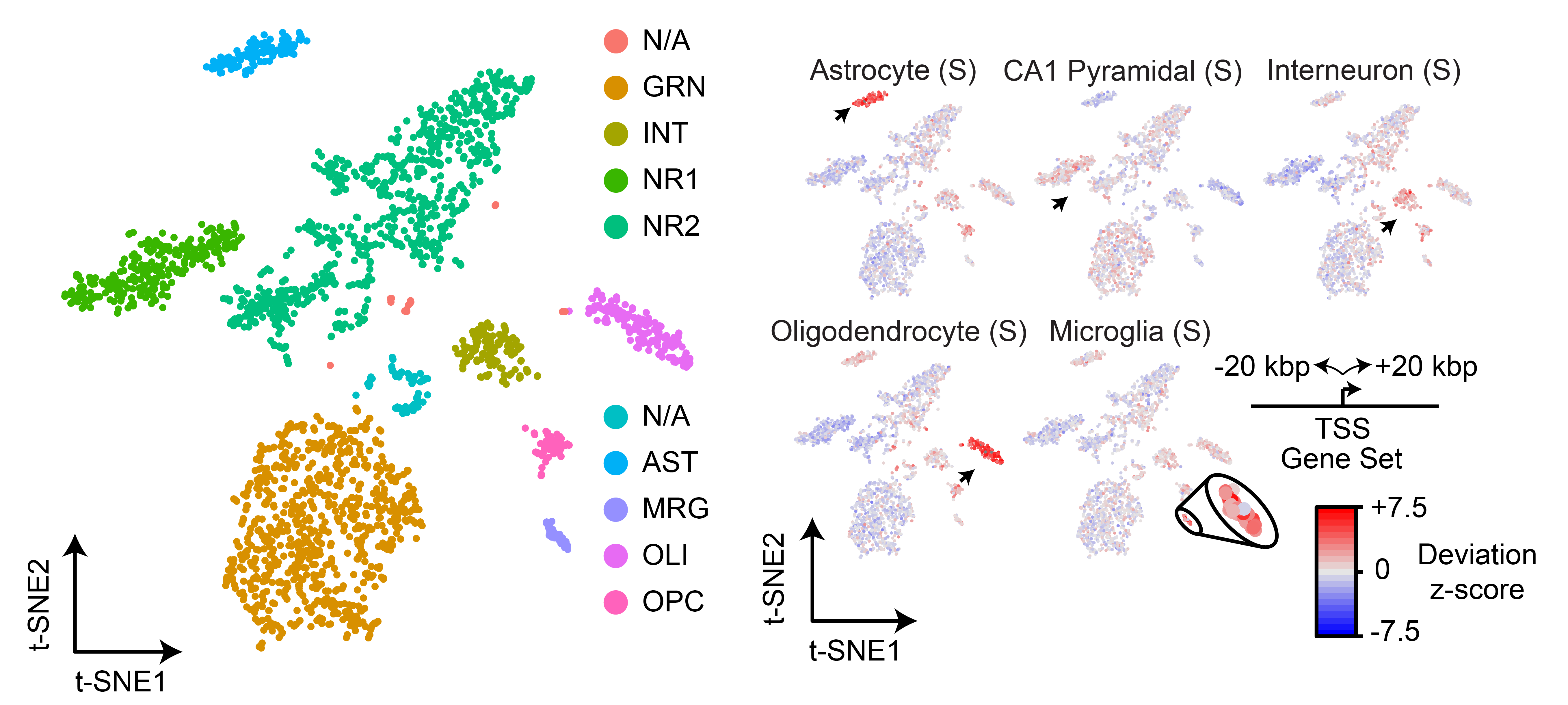
9/4/2018 – New preprint is out on our work profiling the murine hippocampus using sci-ATAC-seq! We are really excited about this work and the datasets which are all available here. We also are releasing ‘scitools’ – a tool we originally developed for internal use for processing and probing single-cell datasets. It is now live here.
8/28/2018 – Dr. Adey is to be recognized as one of the Health Care “Five Under Forty” by the Portland Business Journal at the upcoming event: Health Care of the Future.
7/9/2019 – Dr. Adey was just announced as the recipient of the 2018 American Society of Human Genetics Early-Career Award! Dr. Adey thanks all of those that have supported his work over the years. All of the accomplishments of the lab are the result of outstanding mentors, incredible support at OHSU, and the dedicated members of the lab who make it all happen. Thank you all!
4/16/2018 – sci-MET was featured on the NIH Director’s Blog, highlighting the scalability of combinatorial indexing for single-cell epigenomics!
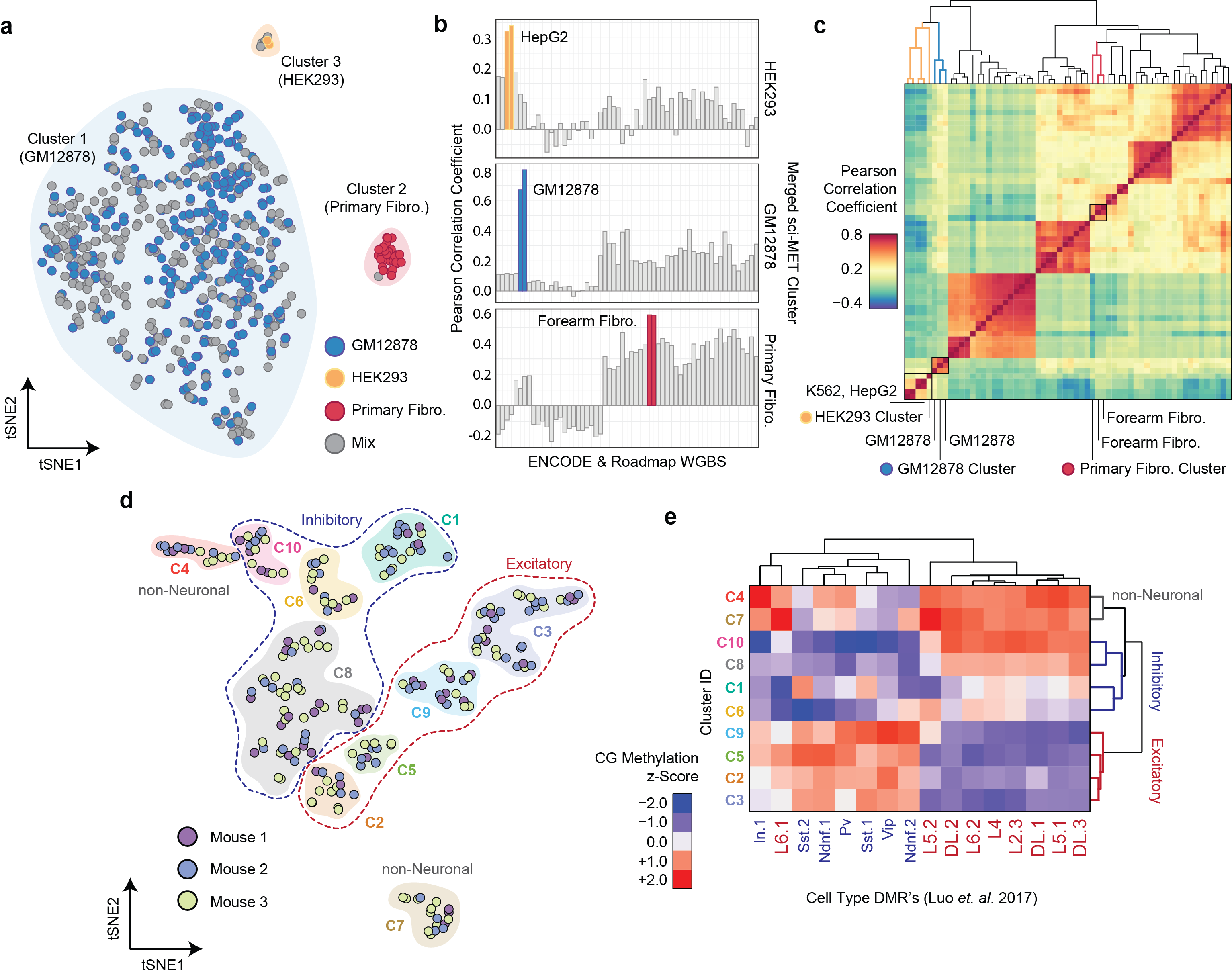
4/9/2018 – New publication out today in Nature Biotechnology on our sci-MET method to profile thousands of single-cells for their DNA methylation profile using our combinatorial indexing platform. Check out figure 2 below, where we demonstrate our ability to discriminate cell types in a mix of human cell lines (top) and from a primary mouse cortex (bottom).
New commentary in PNAS on a single-cell haplotype resolution method out of Kun Zhang’s group.
Dr. Adey was recently interviewed by Nature which appeared in the recent issue focused on single cell biology. Read the interview here.
Published! “Scalable and efficient single-cell DNA methylation sequencing with combinatorial indexing.” (Nature Biotechnology)
In the Adey Lab we seek to answer fundamental questions in biology pertaining to cell identity and cell fate specification. Specifically, we aim to apply systems-based logic to understand the most basic circuitry of a cell and how the configuration of that circuitry locks it in place in its functional role, or pushes it along from one state to another. To accomplish this task, we are leveraging and developing cutting edge technologies to provide the unprecedented resolution and throughput that is required. This includes several single cell strategies to profile genetic and epigenetic properties at high throughput such as single cell ATAC-seq, whole genome sequencing, and DNA Methylation, other important marks that define the epigenetic landscape.